Chemical Evolution
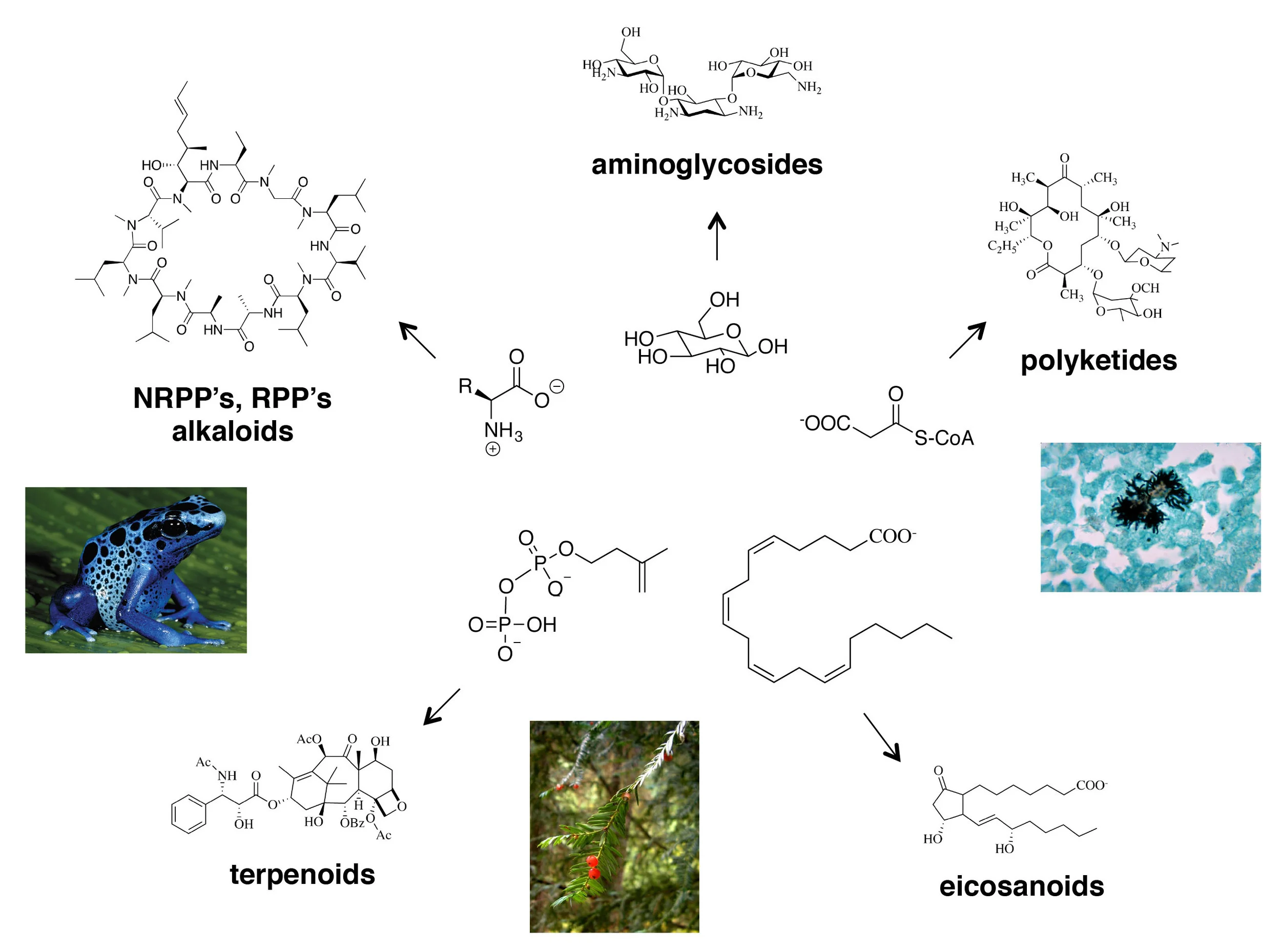
Natural products arose by a process of bio‑combinatorial chemistry, with core metabolic intermediates as the building blocks. In counterclockwise order: (i) Aminoglycosides such as kanamycin derive from basic sugars, (ii) alkaloids and polyketides such as cyclosporin derive from amino acids, (ii) terpenoids such as taxol derive from isopentyl pyrophosphate, the basic building block of steroids, (iv) eicosanoids such as alprostadil derive from arachidonic acid, a lipid component of plasma membranes, (v) polyketides such as erythromycin derive from malonyl-CoA, the basic building block of fatty acids.
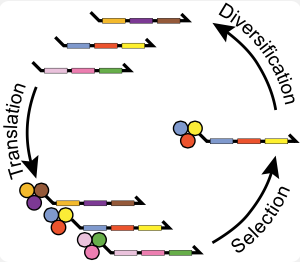
DNA-encoded small molecules can be evolved through multiple generations of translation, selective pressure, amplification and diversification to have unprecedented functionality.
Over half of all medicines derive from natural products, small molecules that evolved over billions of years to control predation, to wage chemical warfare and for cellular communication (examples include taxol and vancomycin; the capsaicin in a steaming bowl of tom yum goong; the jasmone in Chanel No5; the indigo in blue jeans). Natural products came into existence through a process of bio combinatorial chemistry, in which evolving sets of enzymes assemble and covalently modify metabolic intermediates to produce a rich diversity of structures. A small subset of those molecules confer a selective advantage to the organisms that produce them, and are inherited through the DNA blueprints that code for the producing enzymes. The small molecules can thus drive the spread of their associated genetic material. Does modern science have something to learn from nature’s very successful approach to small-molecule discovery? Can we harness its incredible power?
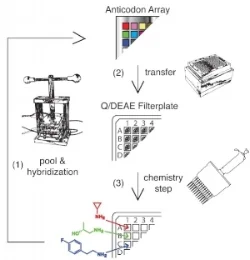
Our lab has worked out an approach to reenact the natural history of chemical evolution in a test tube. Starting with billions of different DNA fragments, we synthesize a small molecule at the end of each fragment such that the small molecule’s structure is programed by the DNA sequence. Each fragment acts like a gene that codes for a chemical reaction sequence. This covalent linkage of small molecules to their genetic material allows us to selectively breed molecular populations. We isolate chemical structures with a desired functional property, and then amplify and diversify the associated jack-pot genes to create a new gene pool. This evolutionary loop is repeated over multiple generations, and the population is then profiled by high-throughput sequencing.
On one hand directed chemical evolution enables fundamental analysis of chemical space. On the other, it lets us make small molecules with tailored properties: specific ligands, substrates and inhibitors of protein targets.
Selected Publications
Directed Chemical Evolution with an Outsized Genetic Code.
Krusemark CJ, Tilmans NP, Brown PO, Harbury PB
PLoS ONE. 11(8): 1-16 (2016)