X-ray Interferometry
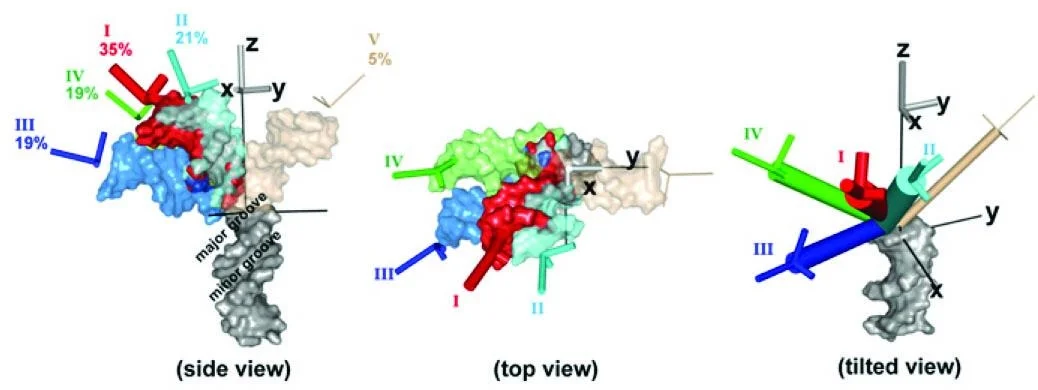
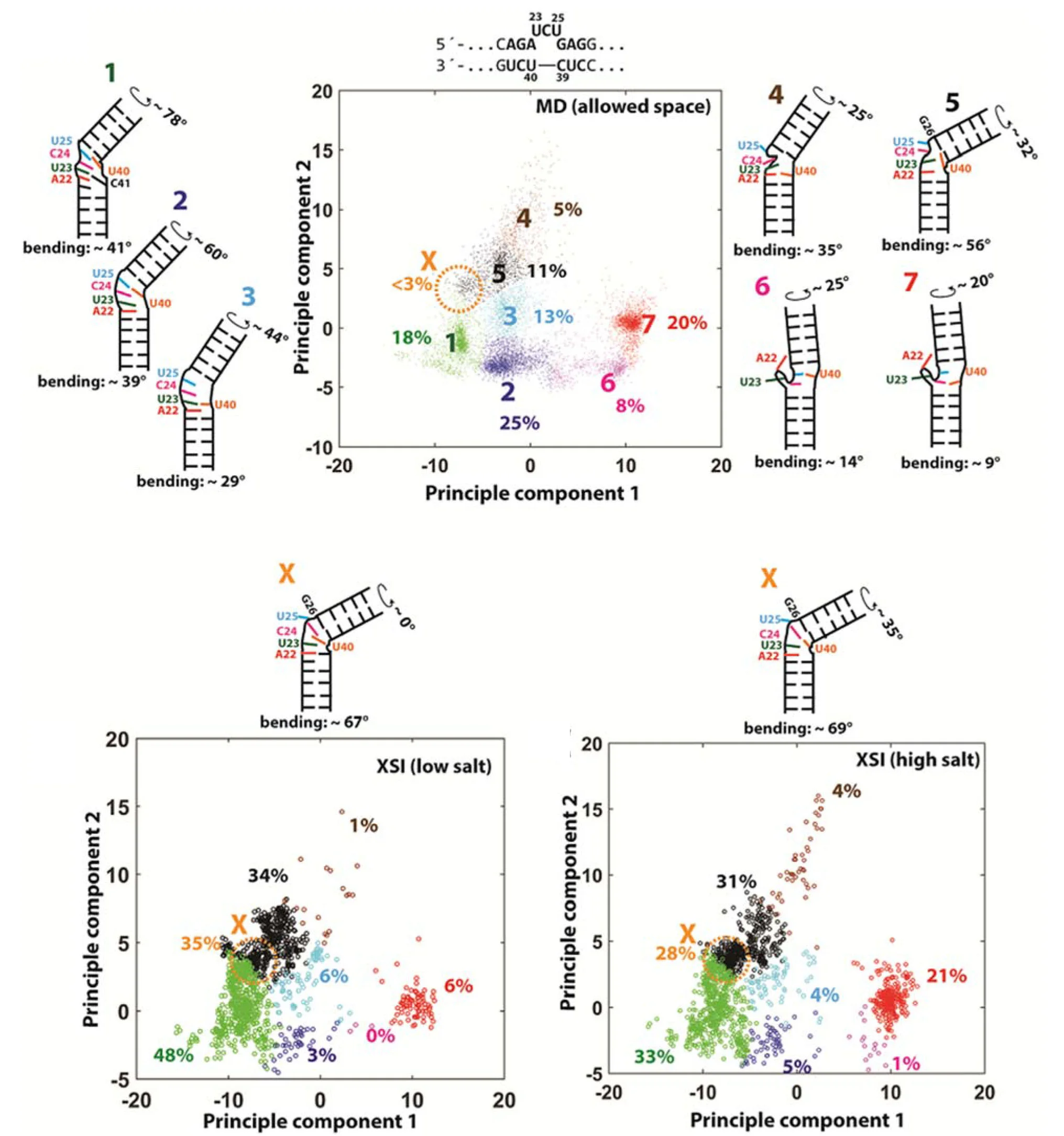
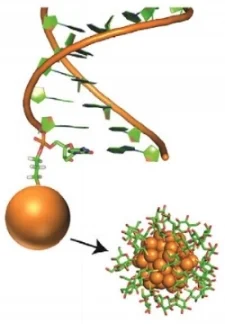
Understanding the ensemble of conformational states adopted by proteins and structured nucleic acids has been a long standing goal of biophysics. At the level of tertiary and quaternary structure, ensembles consist of domain motions and mechanical deformations of a macromolecule. This kind of conformational change is often linked to biological function, for example the structural rearrangements of the ribosome during translation initiation. Determining the three‑dimensional structure of ensembles has been extremely challenging, because the ensembles are heterogeneous and involve nanometer‑scale movements. To address this problem, our lab developed an approach for measuring precise intra-molecular distance distributions in aqueous solution, matching the spatial accuracy of X-ray crystallography. We have used this tool to quantify the microscopic elastic properties of DNA and to determine the three‑dimensional ensemble structure of several ubiquitous nucleic acid motifs, including DNA bulges, RNA kink turns and the TAR RNA (collaboration with the lab of Dan Herschlag). Importantly, we are using the data to develop, test and critique molecular dynamics simulations and coarse‑grained computational models. Ongoing work addresses the geometry and mechanical stiffness of engineered nucleic‑acid nanostructures, and accurate protein structure determination in solution from small‑angle x‑ray scattering measurements.