Genomic Imaging
The human genome encodes a master program for construction of the human body. The program includes subroutines for thousands of different cell types and cell states. Each subroutine prescribes a molecular logic for maintenance of a state, or transition to a new state, depending on local input and output signals. One analogy is Conway's game of life, where two states and four transition rules create a dazzling collection of 'living' creatures. At a high level, the set of cell states and the transition rules in human tissues constitute a complete description of the human program. Knowing them would allow us to understand the meaning of the genome, beyond reading its letters and words. A current challenge is to acquire this knowledge in a comprehensive and efficient way.
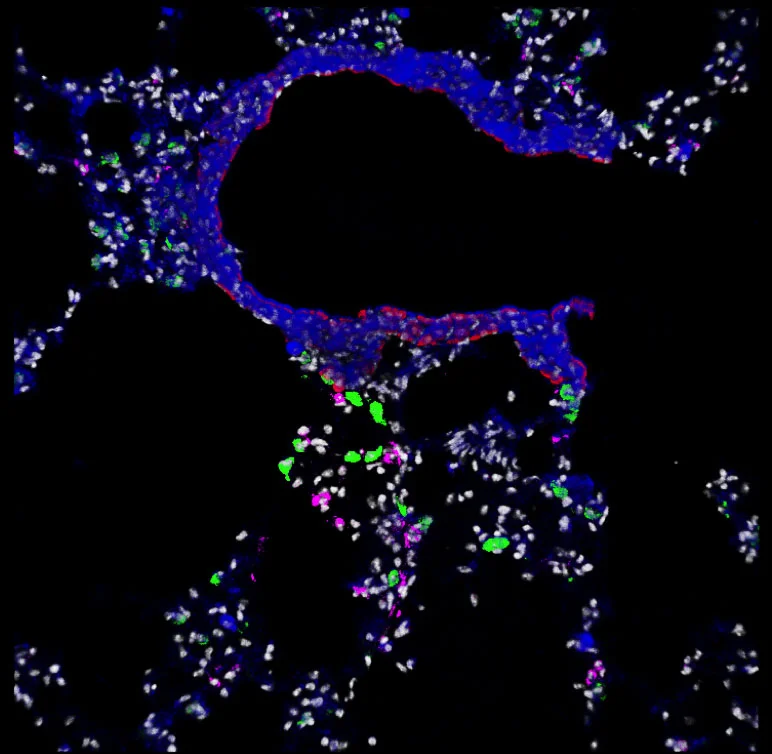
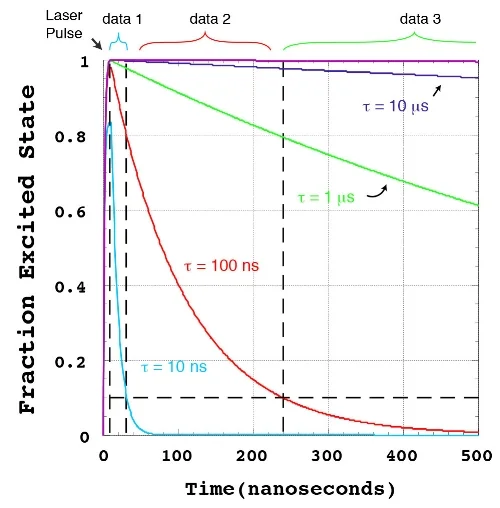
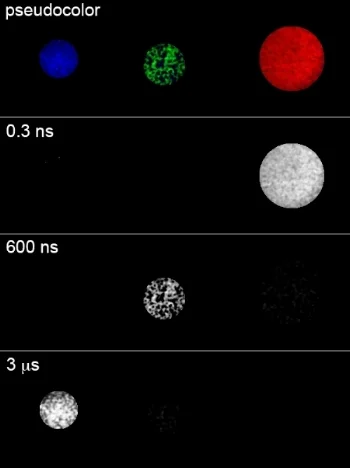
Our aspiration is to empirically learn the human program by genomic imaging. Specifically, we want to simultaneously measure the gene expression patterns of all of the individual cells within an intact three‑dimensional tissue (revealing the subset of genomic instructions that each cell is executing) as well as the signals that are being exchanged between the cells. With Tushar Desai's and Mark Krasnow's labs, we have established scalable techniques for uniquely barcoding the different gene products in tissues. With James Chen's lab, we are working on an optical imaging approach with more than a dozen spectro‑temporal "color channels", so that we can detect many barcodes in a single micrograph. Our aim is to achieve serial imaging in tissues with a rate of information throughput that is comparable to next‑gen sequencing.
Using genomic imaging, we intend to build agent‑based models of the programs that orchestrate human tissue formation. Further, we aim to test the predictions of the models in genetically‑engineered embryoid bodies and in mice. The project presents many fascinating challenges. These range from designing novel fluorophores, constructing microscopes, inventing in situ molecular biology techniques, applying computational inference tools to enormous imaging data sets, and working with experimental systems for analyzing vertebrate development.